In this article, we’ll work through the construction of our own custom Estimator in Pyspark. Estimators act on an input Dataframe, to yield a Pyspark Transformer. They form a key component to Pipelines, where a series of Estimators and Transformers can be chained together to yield a complete machine learning workflow.
Table of Contents

Build a Custom Estimator in Pyspark with 1 Easy Example – image by author
What is a Estimator in Pyspark?
As mentioned in my article on Pyspark Pipelines, a Pyspark Estimator is a class that takes in a Dataframe, and returns a Transformer. In effect, instances of an Estimator are able to learn interesting statistics from the input Dataframe. Once training is done, the learned statistical parameters are then passed to the output Transformer for use in generating predictions. These classes are required to have a fit method.
A high-level overview, of how an Estimator works, is illustrated in Figure 1:
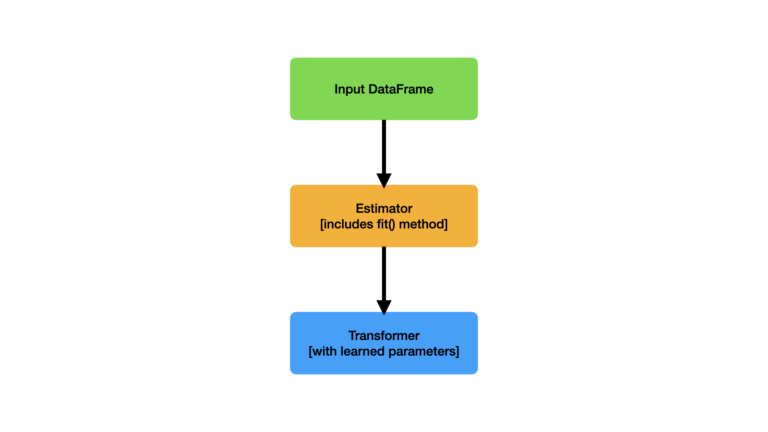
Figure 1: Illustration of an Estimator, and its inputs and outputs. The input Dataframe is passed in through the fit() method, and the output Transformer is returned.
The use of Estimators helps to create modular, reusable code. When defining our own Estimators, there are a couple of points to consider:
- The class must inherit the Pyspark Estimator abstract class
- If you want the ability to save/load your Estimator, the class should also inherit the DefaultParamsReadable and DefaultParamWritable Pyspark classes
We will get into more details as we work through an example. So without any further delay, let’s jump into the code!
Custom Estimator in Pyspark Example
Let’s now work through the implementation of a custom Pyspark Estimator. We will incorporate this Estimator into a pipeline to complete a modeling job.
The aim of this pipeline will be to make use of a Linear Regression model to predict housing values. We will make use of the California Housing Prices dataset. These data were briefly explored in a previous article, and as such I won’t repeat that analysis here.
We can start by importing the necessary packages, and setting up our spark session:
import findspark
findspark.init("/opt/homebrew/Cellar/apache-spark/3.5.0/libexec")
from sklearn.datasets import fetch_california_housing
from typing import Any
import matplotlib.pyplot as plt
from pyspark.sql import SparkSession
from pyspark import keyword_only
from pyspark.sql import functions as F
from pyspark.sql import dataframe as DataFrame
from pyspark.ml import Estimator, Transformer, Pipeline
from pyspark.ml.pipeline import PipelineModel
from pyspark.ml.param.shared import HasInputCols, HasOutputCols, Param, Params, TypeConverters
from pyspark.ml.util import DefaultParamsReadable, DefaultParamsWritable
from pyspark.ml.feature import VectorAssembler
from pyspark.ml.regression import LinearRegression
spark = SparkSession.builder.appName("test").getOrCreate()Dataset
Now let’s obtain the data we will work with:
# obtain data
data = fetch_california_housing(as_frame=True)
dfX = data["data"]
sY = data["target"]
dfX.head(5)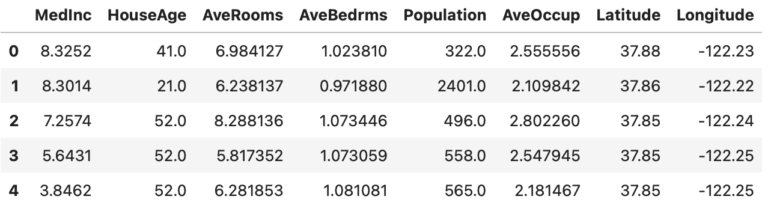
Let’s generate a box plot to visualize the distribution and scale for our input features:
dfX.boxplot(column=dfX.columns.tolist(), rot=45)
plt.xlabel("feature")
plt.ylabel("amount")
plt.show()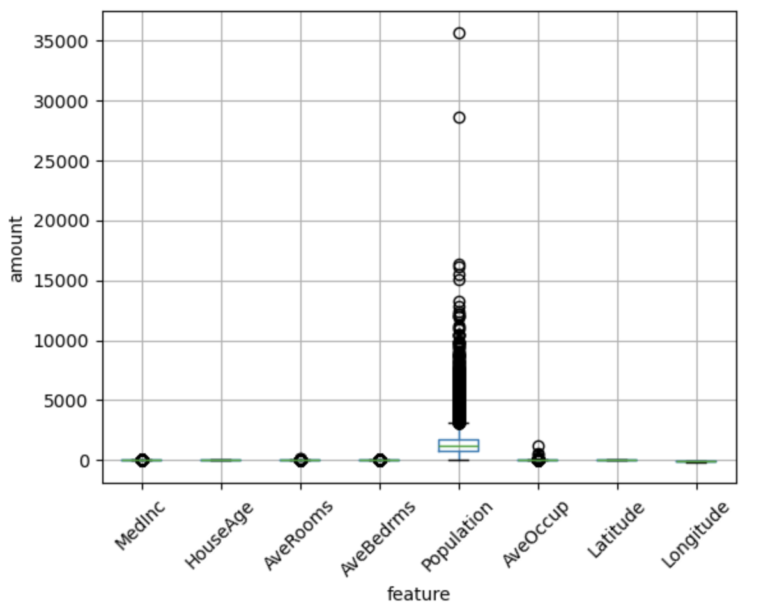
Since we’ll be using Linear Regression, these features should be standardized. Our task in this notebook will be to create an Estimator to do just this! Our Estimator will learn the parameters necessary to convert each input feature to have a mean of 0.0, and standard deviation of 1.0.
Before proceeding, let’s place our data into a Pyspark Dataframe:
dfX["MedHouseVal"] = sY
df = spark.createDataFrame(dfX)Custom Estimator
Now that we know what needs to be done, let’s proceed in a two-step fashion:
- First, we need to build the Transformer that will be returned by the Estimator
- Second, we will implement the Estimator to learn the correct parameters to perform standardization
As such, let’s now start with the implementation of the Transformer to be returned by our Estimator. We can call this Transformer CustomStandardScalerModel:
class CustomStandardScalerModel(Transformer, HasInputCols, HasOutputCols, DefaultParamsReadable, DefaultParamsWritable):
"""
Class to apply learned parameters on a set of input features, in order to standardize them
"""
means = Param(
Params._dummy(),
"means",
"List of mean values for each input feature.",
typeConverter=TypeConverters.toList,
)
stdevs = Param(
Params._dummy(),
"stdevs",
"List of standard deviations for each input feature.",
typeConverter=TypeConverters.toList,
)
@keyword_only
def __init__(self,
inputCols: list=[],
outputCols: list=[],
means: list=[],
stdevs: list=[]) -> None:
"""
Function to initialise the class instance
"""
super().__init__()
self._setDefault(inputCols=[], outputCols=[], means=[], stdevs=[])
kwargs = self._input_kwargs
self.set_params(**kwargs)
# check the input lists have the same length
if len(set([len(inputCols), len(outputCols), len(means), len(stdevs)])) != 1:
raise ValueError("Input lists have non-matching length!")
@keyword_only
def set_params(self,
inputCols: list=[],
outputCols: list=[],
means: list=[],
stdevs: list=[]) -> None:
kwargs = self._input_kwargs
self._set(**kwargs)
def setMeans(self, new_means: list) -> Any:
return self.setParams(means=new_means)
def getMeans(self) -> list:
return self.getOrDefault(self.means)
def setStdevs(self, new_stdevs: list) -> Any:
return self.setParams(stdevs=new_stdevs)
def getStdevs(self) -> list:
return self.getOrDefault(self.stdevs)
def _transform(self, df: DataFrame) -> DataFrame:
"""
Function to standardize a set of input features in a dataframe
"""
# read in the transformer parameters
inputCols = self.getInputCols()
outputCols = self.getOutputCols()
means = self.getMeans()
stdevs = self.getStdevs()
# generate standardized output columns, and return
for input_col, output_col, mean, stdev in zip(inputCols, outputCols, means, stdevs):
df = df.withColumn(output_col, (F.col(input_col) - mean)/stdev)
return dfThere are a few interesting things to note with this section of code:
- In addition to inheriting Transformer, DefaultParamsReadable, and DefaultParamsWritable, this class is also inherits HasInputCols and HasOutputCols. These Mixins are used to define a list of input and output columns.
- When we want to create an input that isn’t defined by a Pyspark Mixin, we will have to define it ourselves. This is the case for the means and stdevs parameters, which will be lists containing the means and standard deviations necessary for standardization. I have written the definition for these parameters immediately before the initialization function (lines 5 to 16).
- The initializer function proceeds in the following way:
- The initializers of the parent classes are called
- Default values for the 4 input arguments are set
- Input keyword arguments are read and set
- The _transform function is where the main action happens. This is where we can define our logic to standardize an input Dataframe. Note that the lists of means and standard deviations are used on each input column, to yield a corresponding output column. This function is executed when the transform method is called on the class instance.
Now that our output is defined, we can now proceed to implement our Estimator CustomStandardScaler:
class CustomStandardScaler(Estimator, HasInputCols, HasOutputCols, DefaultParamsReadable, DefaultParamsWritable):
"""
Class to learn parameters needed to standardize a set of input features
"""
withMean = Param(
Params._dummy(),
"withMean",
"Boolean to determine if standardizing will involve the mean.",
typeConverter=TypeConverters.toBoolean,
)
withStd = Param(
Params._dummy(),
"withStd",
"Boolean to determine if standardizing will involve the standard deviation.",
typeConverter=TypeConverters.toBoolean,
)
@keyword_only
def __init__(self,
inputCols: list=[],
outputCols: list=[],
withMean: bool=True,
withStd: bool=True) -> None:
"""
Function to initialise the class instance
"""
super().__init__()
self._setDefault(inputCols=[], outputCols=[], withMean=True, withStd=True)
kwargs = self._input_kwargs
self.set_params(**kwargs)
# check the input lists have the same length
if len(inputCols) != len(outputCols):
raise ValueError("Input and output columns have non-matching length!")
@keyword_only
def set_params(self,
inputCols: list=[],
outputCols: list=[],
withMean: bool=True,
withStd: bool=True) -> None:
kwargs = self._input_kwargs
self._set(**kwargs)
def setWithMean(self, new_withMean: bool) -> Any:
return self.setParams(withMean=new_withMean)
def getWithMean(self) -> bool:
return self.getOrDefault(self.withMean)
def setWithStd(self, new_withStd: bool) -> Any:
return self.setParams(withStd=new_withStd)
def getWithStd(self) -> bool:
return self.getOrDefault(self.withStd)
def _fit(self, df: DataFrame) -> CustomStandardScalerModel:
"""
Function to learn the standardizing parameters
"""
# read in the estimator parameters
inputCols = self.getInputCols()
outputCols = self.getOutputCols()
withMean = self.getWithMean()
withStd = self.getWithStd()
# compute means
if withMean:
means = [df.select(F.mean(col)).collect()[0][0] for col in inputCols]
else:
means = [0.0 for col in inputCols]
# compute standard deviations
if withStd:
stdevs = [df.select(F.std(col)).collect()[0][0] for col in inputCols]
else:
stdevs = [1.0 for col in inputCols]
# return transformer
return CustomStandardScalerModel(inputCols=inputCols, outputCols=outputCols, means=means, stdevs=stdevs)Some things to note with this implementation:
- The inheritance, parameter setup, and initialiser are similar to the Transformer case, except we inherit the Estimator abstract class.
- The input parameters withMean and withStd are booleans, meant to control how the standardizing will be carried out.
- The real logic happens in the _fit function, where we specify how the parameters (means and standard deviations in this case) will be learned. Afterwards, an instance of CustomStandardScalerModel is returned. This function is executed when fit is called on a class instance.
Testing the Custom Estimator
Now that the implementation is done, we can test out our Estimator to verify that it works. Let’s create an instance, and then pass our data through it:
# prepare input columns list
input_columns = df.columns
input_columns.remove("MedianHouseValue")
# prepare output columns list
output_columns = [col + "_s" for col in input_columns]
scalar = CustomStandardScaler(inputCols=input_columns, outputCols=output_columns, withMean=True, withStd=True)
scalar_model = scalar.fit(df)We can do a quick check to see the values of the means and standard deviations for each input column:
# check our learned means
scalar_model.getMeans()[3.8706710029069753, 28.639486434108527, 5.428999742190377, 1.0966751496062066, 1425.4767441860465, 3.070655159436372, 35.6318614341085, -119.56970445736444]
# check our learned standard deviations
scalar_model.getStdevs()[1.899821717945268, 12.585557612111646, 2.474173139424319, 0.4739108567954659, 1132.4621217653414, 10.386049562213614, 2.135952397457123, 2.0035317235025847]
These values look good. Now let’s transform our data and visualize the results:
df = scalar_model.transform(df)
# plot the standardized features
dfP = df.select(output_columns).toPandas()
ax = dfP.boxplot(column=dfP.columns.tolist(), rot=45)
ax.set_ylim(-5, 5)
plt.xlabel("feature")
plt.ylabel("amount")
plt.show()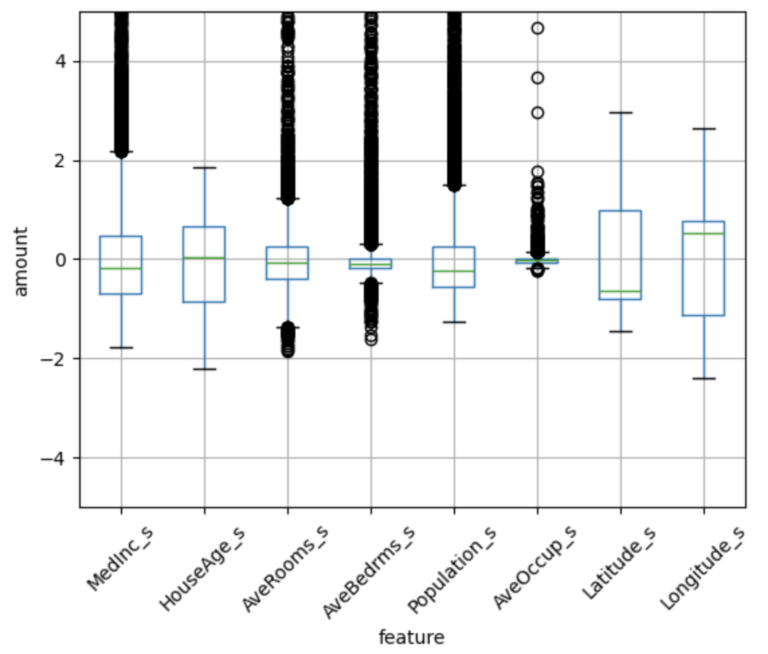
We can see that our custom standard scaler has transformed our data, such that all the features exist at similar scales. It is important to note that differences do persist between the features, however (i.e. number and distribution of outliers, range of quartiles, etc). Nevertheless, these results are exactly what we would expect from standardization (refer to this previous article).
Using our Estimator in a Pipeline
The strength of writing our own Estimators comes when applying them within a machine learning pipeline. Here we’ll now incorporate an instance of CustomStandardScaler into a pipeline to model our data.
Let’s start by doing a train-test split on the available data:
# perform a train-test split
dfTrain, dfTest = df.randomSplit([0.8,0.2])As mentioned previously, I’ll be making use of a Linear Regression to model these data. Now we can implement a pipeline, train it, and then view the results on the test set:
# define the pipeline
pipeline = Pipeline(stages=[
CustomStandardScaler(inputCols=input_columns, outputCols=output_columns),
VectorAssembler(inputCols = output_columns, outputCol="Features"),
LinearRegression(featuresCol = 'Features', labelCol = 'MedianHouseValue', regParam = 0.01)
])
# train the pipeline
model = pipeline.fit(dfTrain)
# make predictions on the test set
dfPred = model.transform(dfTest)
dfPred.select("MedianHouseValue","prediction").show(5)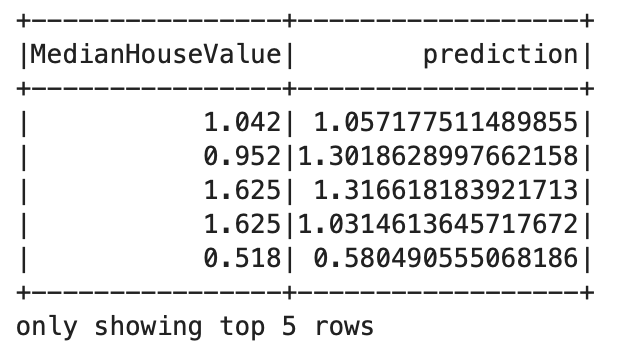
It is evident that the pipeline model is working as expected.
Another nice benefit of pipelines is that we can save our entire trained workflow, for use at a later time. Let’s try to save our pipeline, load it, and make use of the loaded instance:
# write the trained model to disk
model.write().overwrite().save("./trained_pipeline")
# load the saved model, and confirm it works
model2 = PipelineModel.load("./trained_pipeline")
dfPred = model2.transform(dfTest)
dfPred.select("MedHouseVal","prediction").show(5)
This result is exactly the same as that obtained above.
Final Remarks
In this post you have learned:
- What a Pyspark Estimator is.
- How to build your own Estimators in Pyspark.
- How to implement a custom Estimator within a Pyspark pipeline.
I hope you enjoyed this article, and gained some value from it. If you would like to take a closer look at the code presented here, please take a look at my GitHub. If you have any questions or suggestions, please feel free to add a comment below. Your input is greatly appreciated.
Note I have started a New Monthly Newsletter! At the end of each month I will send out this free newsletter to each of my subscribers by email. This is the best way to stay on top of my latest content. Sign up for the newsletter here!
Related Posts
Hi I'm Michael Attard, a Data Scientist with a background in Astrophysics. I enjoy helping others on their journey to learn more about machine learning, and how it can be applied in industry.
